The devil is in the detail
I was learning some steganography methods for a security project. At the same time, I was writing up a paper with some friends (https://link.springer.com/content/pdf/11.1007%2Fs10875-019-00670-z.pdf) and decided to hide a little message in one of the figures. The goal was to encode some hidden data and see could we get it back out the other side, after publication.
It would be a lot of trouble for a master of espionage to have to publish a paper every time a secret message needed to be encoded for public transmission, but maybe it could be done!
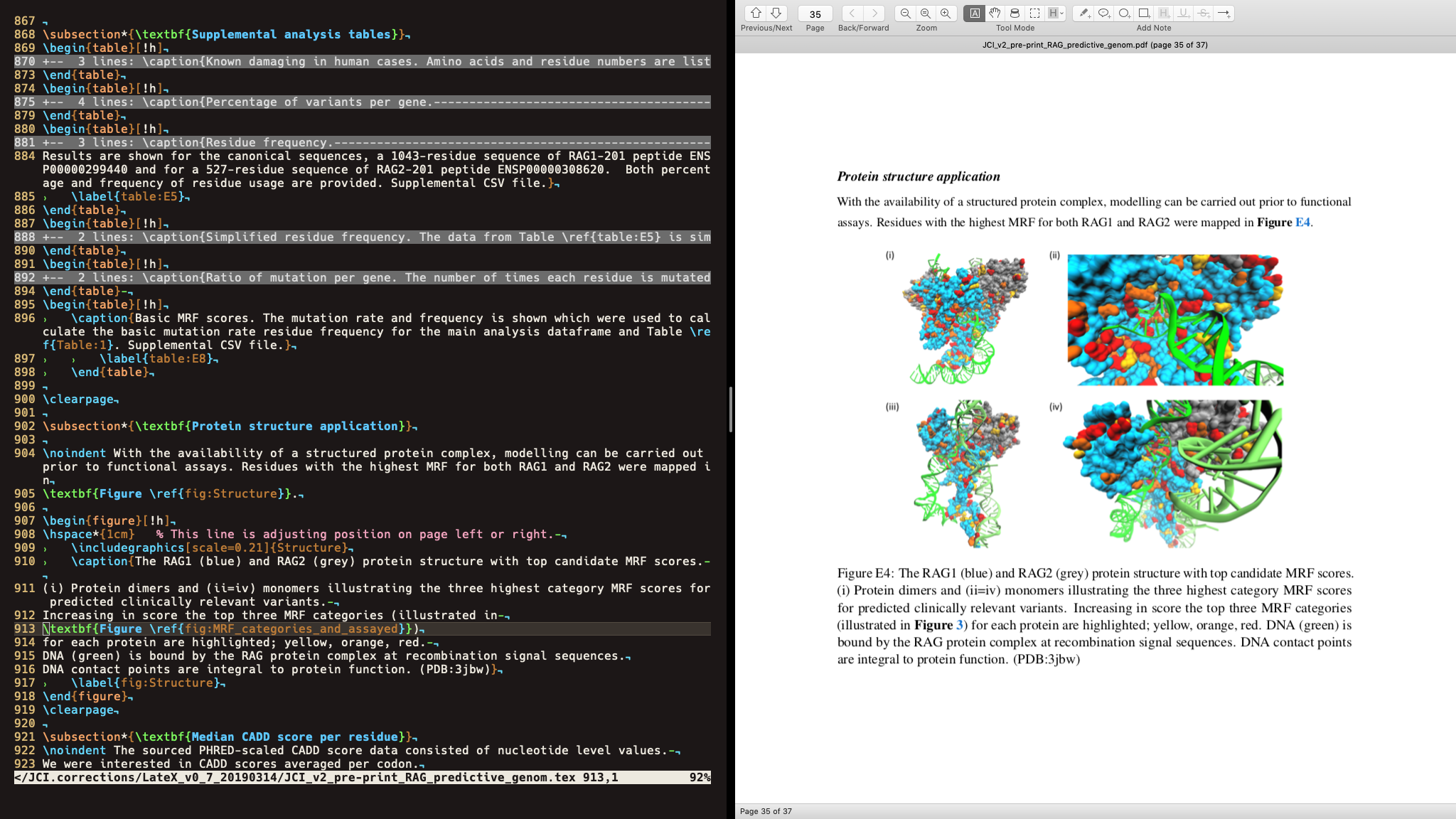
The secret message was imported into R from plain text. It was encoded into a png file that was to be a supplemental figure in our paper From experience, supplemental figures usually get uploaded to a repository and stored in the same format for later downloaded after publication, until main-text figures which are often converted in one way or another by the publisher. Once the figure was released, anyone with the correct decoding algorithm could get back the secret message.
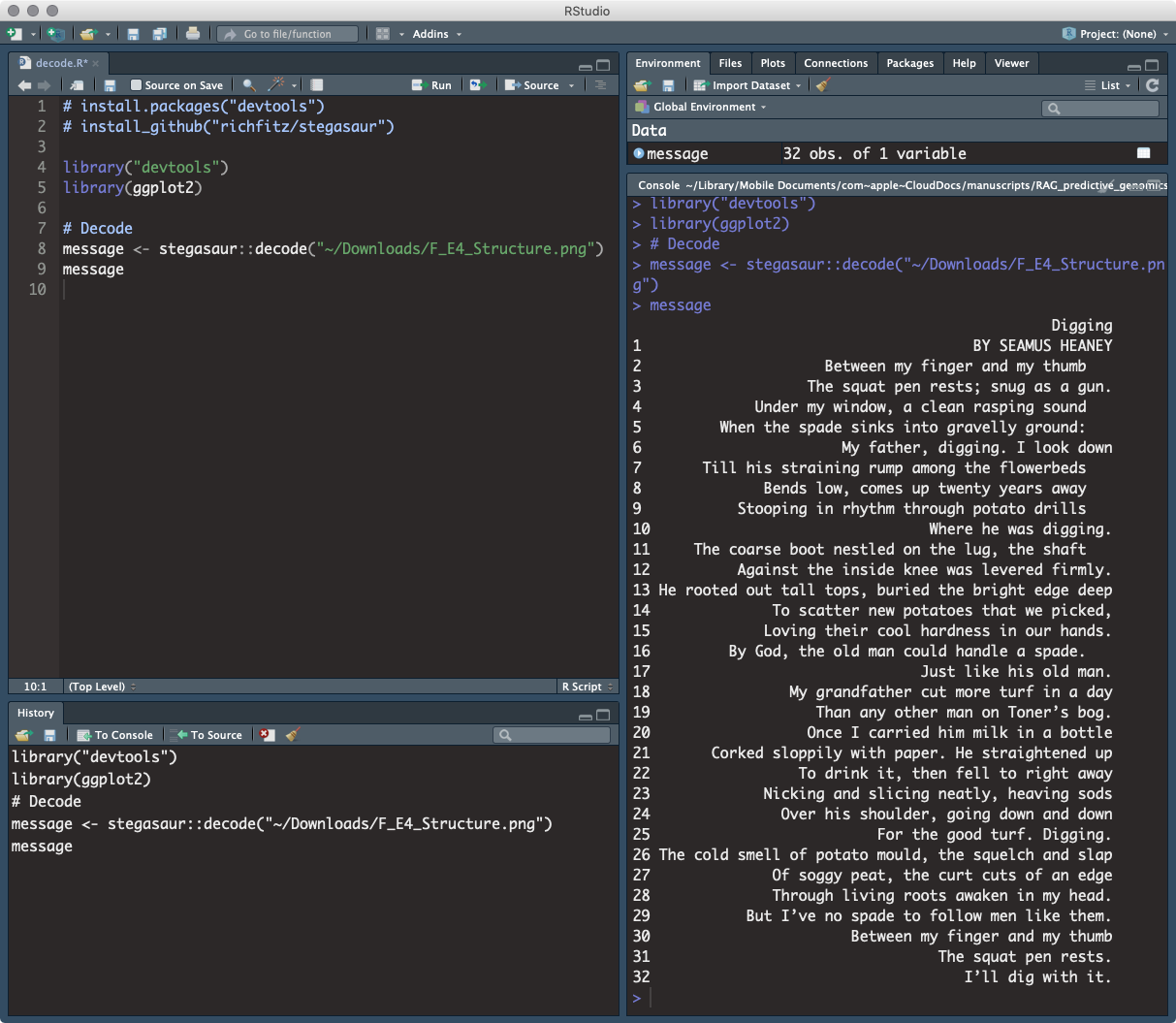
In this case, I used the richfitz/stegasaur package, innocently offered with “The aim is to be able to encode arbitrary R objects in cat pictures.” The inforation was hidden within the data that makes up the image, and is imperceivable to the naked eye. Even better, the file size was slightly SMALLER than the original (1.9MB to 1.7MB) without a noticeable difference in quality. After peer review of the paper, publisher proceessing and publication, the secret message was successfully decoded from the protein structure figure “F_E4_Structure.png” and was printed to the console:
This exercise looks like a bit of fun, but was done to demonstrate the unavoidable risks that exist in sensitive data research.1 Today, protected data enclaves are used for processing sensitive data and usually allow summary results or figures to be exported. These are therefore basically a trust based system and are otherwise only security theatre.
Here’s an idea, steganographic message blocks seperated across a career worth of publications, that can only be deciphered once all blocks are collected! Who would go to that trouble? Go see what you can find in one Supplementary material png file. https://link.springer.com/article/10.1007%2Fs10875-019-00670-z
R script
Try it yourself. Find the paper and get Figure E4. Download 10875_2019_670_MOESM5_ESM
> install.packages("devtools")
> install_github("richfitz/stegasaur")
> library("devtools")
> library(ggplot2)
> Decode
message <-
> stegasaur::decode("~/10875_2019_670_MOESM5_ESM.png")