SARS-CoV-2 variants of concern
Variants of concern aligned to vaccine coding sequences
This work has not been peer reviewed.
Open PDF visualisation - variants_of_concern_to_vaccine.pdf
Aims and results
- To produce standardized alignments of vaccine sequences.
- Determine which are at risk to emerging variants.
From the data presented within:
- The translated amino acid sequences for all vaccines were derived.
- These were aligned to the SARS-CoV-2 reference amino acid sequences of spike glycoprotein.
- Known variants-of-concern were then annotated and visualised.
Overview
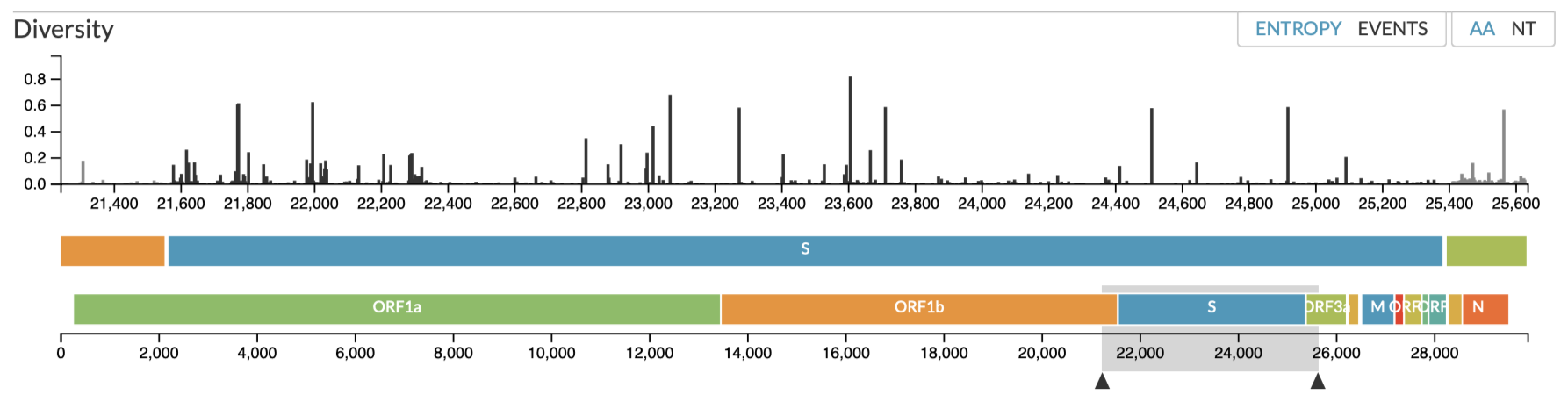
Variants-of-concern for five SARS-CoV-2 strains (CDC 4 Jun 2021) are illustrated against the translated amino acid sequences of the vaccines;
- Moderna mRNA-1273
- Pfizer/BioNTech BNT-162b2
- Janssen/Johnson & Johnson Ad26.COV2-S
- Novavax NVX-CoV2373
- Curevac CVnCoV
- Sputnik V
- AstraZeneca AZD1222
and reference genome sequences;
- QHD43416.1 [MN908947.3] and
- YP_009724390.1 [NC_045512.2].
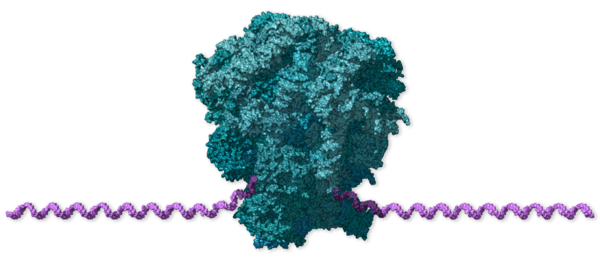
The variants-of-concern are shown here, illustrated on the protein structure; 6ZOX.pdb DOI: 10.2210/pdb6ZOX/pdb Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x2 disulphide-bond mutant, G413C, V987C, single Arg S1/S2 cleavage site), provided by Xiong et al., 2020 10.1038/s41594-020-0478-5.

Two of the defining genetic features that are different between vaccines are seen here,
- the S glycoprotein furin cleavage modification region (p.682-685)

- the S glycoprotein stabilization modification region (p.986-987)
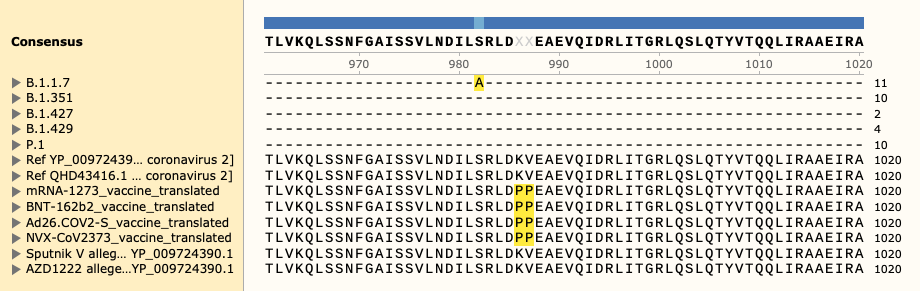
Visual alignment is shown against translated coding sequence for spike glycoprotein, illustrated here via
Fasta sequences are included for:
- Variants of Concern B.1.1.7
- Variants of Concern B.1.351
- Variants of Concern B.1.427
- Variants of Concern B.1.429
- Variants of Concern P.1
- Ref QHD43416.1 [MN908947.3]
- Ref YP_009724390.1 [NC_045512.2]
- mRNA-1273 vaccine translated
- BNT-162b2 vaccine translated
- Ad26.COV2-S vaccine translated
- NVX-CoV2373 vaccine translated
- Sputnik V alleged unmodified YP_009724390.1
- AZD1222 alleged unmodified YP_009724390.1
Reference genome sequence
The two reference sequences that are used by vaccine developers are;
- Complete genome DNA and translated coding sequences:
- NC_045512.2,
- Protein ID for spike glycoprotein YP_009724390.1.
- Date: 18-JUL-2020
- This the same translated protein sequence as that is referred to based on genomic coordinates NC_045512.2:21563-25384 translated GU280_gp02
- Complete genome DNA and translated coding sequences:
- MN908947.3
- Protein ID for spike glycoprotein QHD43416.1
- Date: 18-MAR-2020
Both reference sequences are provided in files:
Vaccine sequence reproduction
The sequences for vaccines have been reproduced by careful reconstruction based on
- The authors’ reported reference sequence and
- The description of the genetic modifications used during vaccine development.
The primary sources are provided in each case, along with a detailed description of the genetic variants provided by authors. Additionally, the correct HGVS-recommended nomenclature has been used for more reliable reproduction than some of the primary sources.
For visual simplicity, an X symbol was used to illustrate amino acid deletions. All other amino acid changes use their correct symbol.
For vaccines BNT-162b2 and mRNA-1273, the assemblies have also been sourced from NAalytics. This data matches the vaccine sequences that have been reproduced here based on primary literature. Briefly, their experimental sequence information from the initial Moderna (Corbett Nature 2020 Oct) and Pfizer/BioNTech (Polack NEJM 2020 Dec) COVID-19 vaccines, allowed them to produce a working assembly of the former and a confirmation of previously reported sequence information for the latter RNA. Their data was sourced and formatted to select the coding sequences. The nucleotide sequences were then translated into amino acid coding sequences using https://web.expasy.org/translate/, as shown in files:
sarscov2_vaccine_sequence_translated_mRNA-1273.md sarscov2_vaccine_sequence_translated_BNT-162b2.md
Covid-19 vaccine sequences summarised
The correct HGVS standard notation is used.
- mRNA-1273
- Genetics: p.(Lys986_Val987delinsProPro) - stabilizing x2 (PP)
- Delivery: Lipid-nanoparticle
- BNT162b2
- Genetics: p.(Lys986_Val987delinsProPro) - stabilizing x2 (PP)
- Delivery: Lipid-nanoparticle
- Ad26.COV2-S
- Genetics: p.[Arg682Ser;p.Arg685Gln] - furin cleavage x2 (SRAG)
- Genetics: p.(Lys986_Val987delinsProPro) - stabilizing x2 (PP)
- Delivery: Adenovirus vector (Ad26)
- NVX-CoV2373
- Genetics: p.[Arg682_Arg683delinsGlnGln;Arg685Gln] - furin cleavage x3 (GGAG)
- Genetics: p.(Lys986_Val987delinsProPro) - stabilizing x2 (PP)
- Delivery: Lipid-nanoparticle, baculovirus expression cultured in Sf9
- Sputnik V
- Genetics: “unmodified” full-length S-protein
- Genetics: No reference sequence found
- Delivery: Adenovirus vectors (Ad26 dose 1) and (Ad5 dose 2)
-
Incomplete others:
- CVnCoV
- Genetics: modified S protein.
- AZD1222
- Genetics: Unmodified S protein
- Genetics: No reference sequence found
- Adenovirus vector (ChAdOx1).
- CoronaVac
- a preparation of inactivated SARS-CoV-2 virions.
Covid-19 vaccine details
- BioNTech/Pfizer: BNT162b2
- Modified mRNA-in-lipid-nanoparticle vaccine
- Expressing a modified S protein.
- Stabiliazation by proline substitutions p.K986P, p.V987P.
- Moderna: mRNA-1273
- Modified mRNA-in-lipid-nanoparticle vaccine
- Expressing a modified S protein.
- Stabiliazation by proline substitutions p.K986P, p.V987P.
- Janssen/Johnson & Johnson: Ad26.COV2-S aka JNJ-78436735
- Pre-prindt, Published.
- Adenovirus serotype 26 (Ad26) viral vector vaccine
- Expressing a modified S protein.
- S protein of SARS-CoV-2 corresponding to positions 21,536–25,384 in SARS-CoV-2 isolate Wuhan-Hu-1 (genome MN908947 (18-MAR-2020))Published.
- For Ad26.S.PP, the two stabilising variants p.(Lys986_Val987delinsProPro) are included as well as two mutations in the furin cleavage site that preserve the prefusion conformation and blocks shedding of S1.
- The furin cleavage site was abolished by amino acid changes p.R682S and p.R685G.
- Stabiliazation by proline substitutions p.K986P, p.V987P.
- The correct HGVS standard notation should be: p.[Arg682Ser;p.Arg685Gln] and p.(Lys986_Val987delinsProPro).
- Novavax: NVX-CoV2373
- A protein subunit vaccine containing a doubly modified S protein, with adjuvant.
- Part of a 27.2nm nanoparticle.
- S protein of SARS-CoV-2 corresponding to GenBank MN908947 nucleotides 21563-25384 as published.
- Contains the modified S protein with the two Proline substitutions, K986P and V987P. Additionally, three amino acids are changed (682-RRAR-685 to 682-QQAQ-685) to protect the protein against proteases.
- The authors failed to write the correct HGVS standard notation: p.[Arg682_Arg683delinsGlnGln;Arg685Gln] and p.(Lys986_Val987delinsProPro), a simple list would even be better: p.R682Q, p.R683Q, p.R685Q, p.K986P, and p.V987P.
- Saponin-based Matrix-M adjuvent.
- Protein expression by a baculovirus in an Sf9 insect infection culture.
- https://www.biorxiv.org/content/10.1101/2020.06.29.178509v1.full.pdf
- Delivery in lipid nanoparticle
- Gamaleya Research Institute of Epidemiology and Microbiology: Sputnik V
- aka Гам-КОВИД-Вак (Gam-COVID-Vac).
- Two differnt adenovirus viral vectors.
- Uses two different adenovirus serotypes; recombinant Ad26 (dose 1) and recombinant Ad5 (dose 2).
- Both carrying the gene for Spike glycoprotein (rAd26-S and rAd5-S).
- Antigen insert is an “unmodified” full-length S-protein (no reference sequence).
- Produced in HEK293 cell line.
- No reference sequence found
- The first major paper [Logunov et al Lancet. 2020] is this clinical trial of frozen and lyophilised vaccine. It mentions previous unpublished pre-clinical trials. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7471804/
- Curevac: CVnCoV
- Unmodified mRNA-in-lipid-nanoparticle vaccine
- Expressing a modified S protein.
- Based on genome from first isolate NC_045512.2, Spike glycoprotein YP_009724390.1.
- full-length S featuring K986P and V987P mutations.
- https://pubmed.ncbi.nlm.nih.gov/33863911/
- Oxford/AstraZeneca: AZD1222 (formerly ChAdOx1 nCoV-19)
- A viral vector vaccine
- Viral vector (ChAdOx1 - chimpanzee adenovirus Oxford 1, as published)
- Expressing the unmodified S protein.
- No reference sequence found
- Folegatti et al, Lancet July 2020
Vaccine Multiple Sequence Alignment
The amino acid sequences of the coding region from each of the vaccine sequences and the reference sequence were used for multiple sequence alignment via https://www.ebi.ac.uk/Tools/msa/clustalo/.
Variants-of-concern were then formatted to be used for annotation on the aligned sequences.
Variants of concern
SARS-CoV-2 Variant Classifications and Definitions were derived from https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Fcases-updates%2Fvariant-surveillance%2Fvariant-info.html
This dataset includes:
- Variants of Interest (VOI)
- Variants of Concern (VOC)
- Variants of High Consequence (VOHC)
The reformatted tables are presented in files:
There are currently no VOHC. VOC (but not VOI) were presented in the final visualisation.
Aligned variants-of-concern to vaccine
The variants of concern were formatted such that one pseudo-fasta format entry contains the amino acid change for each strain. This data was then added to the multiple sequence alignment file to allow for aligned annotations, as shown in file:
variants_of_concern_to_vaccine.fa
The file contains the list the variants-of-concern for five Sars-CoV-2 strains, 2 reference sequence, and 6 vaccine sequences:
- Variants of Concern B.1.1.7
- Variants of Concern B.1.351
- Variants of Concern B.1.427
- Variants of Concern B.1.429
- Variants of Concern P.1
- Ref QHD43416.1 [MN908947.3]
- Ref YP_009724390.1 [NC_045512.2]
- mRNA-1273 vaccine translated
- BNT-162b2 vaccine translated
- Ad26.COV2-S vaccine translated
- NVX-CoV2373 vaccine translated
- Sputnik V alleged unmodified YP_009724390.1
- AZD1222 alleged unmodified YP_009724390.1
Different strains will contain benign variants. Typically, full sequences are used for alignment. However, this can be visually distracting. Instead, only the variants-of-concern are annotated for the strain sequences.
The final illustration was made using https://www.snapgene.com software. The snapgene-software formatted output can be loaded with the file:
variants_of_concern_to_vaccine.praln
The final PDF version is shown in file: Open PDF visualisation variants_of_concern_to_vaccine.pdf
Main files
The main files that might interst you are listed here together. Other files that are not listed contain intermediate data.
- reference_sequence_NC_045512.2.21563-25384.md
- sarscov2_vaccine_sequence_translated_mRNA-1273.md
- sarscov2_vaccine_sequence_translated_BNT-162b2.md
- sarscov2_vaccine_sequence_translated_NVX-CoV2373.md
- sarscov2_vaccine_sequence_translated_Ad26.COV2-S.md
- sarscov2_vaccine_sequence_translated_AZD1222.md
- sarscov2_vaccine_sequence_translated_NVX-CoV2373.md
- variants_of_concern.xlsx
- variants_of_concern.csv
- variants_of_concern_to_vaccine.fa
- variants_of_concern_to_vaccine.praln
- Protein structure, original structure file
- Protein structure, VMD visualisation state 6zox_voc_20210604_stage.vmd
- Protein structure with variants of concern 6zox_voc_20210604_front.jpg
- Protein structure with variants of concern 6zox_voc_20210604_side.jpg
- Protein structure with variants of concern 6zox_voc_20210604_top.jpg
Other notes