Multiple transcript variant
Introduction
A colleague asked me about why there are multiple mRNA transcripts with different variant positions from a single variant in human genomic DNA. Here is a short summary to answer that question (forgiving over-simplifications). First, a quick recap of how DNA is used to produce proteins. Then an example of a gene where the mRNA could be spliced more than one way by the splisosome complex.
DNA to protein
- Genomic DNA (only one version per cell/individual).
- Transcription to pre-mRNA (one exact copy of DNA as mRNA).
- Splicing pre-mRNA to mRNA transcript (introns removed by splisosome). One version or several slightly different versions.
- Spliced mRNA (messenger mRNA) translated into amino acid peptide chain.
- Peptide chain folds into protein.
Genomic DNA is transcribed over and over, in every cell (ignoring tissue-specific expression, dynamics, etc.). When the pre-mRNA is spliced, there may be multiple versions produced. It all depends on how the splicing complex recognizes the intron/exon sequences. This also explains why splice mutations occur in 4-5 the nucleotides around the junction between exon and intron. The is the part of the sequence that is recognised by the splisosome.
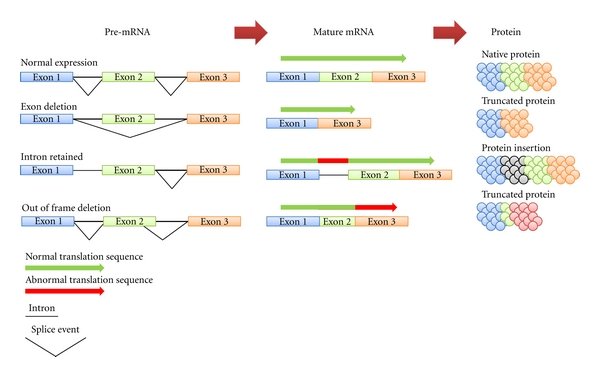
Figure 1: Transcripts from a single gene undergo different splicing events. This figure illustrate why one single variant in genomic DNA can result in several different variants in the RNA transcript / protein isoforms. E.g. a variant in exon 2 will be present in 3 of the 4 protein isoforms and its amino acid position (nomenclature) will vary. Note for simplicity when reporting a variant, we often report just the genomic position or the canonical transcript position (in cDNA) instead of reporting all possible outcomes (description of cDNA below). HGVS has guidelines on nomenclature - varnomen.hgvs.org.
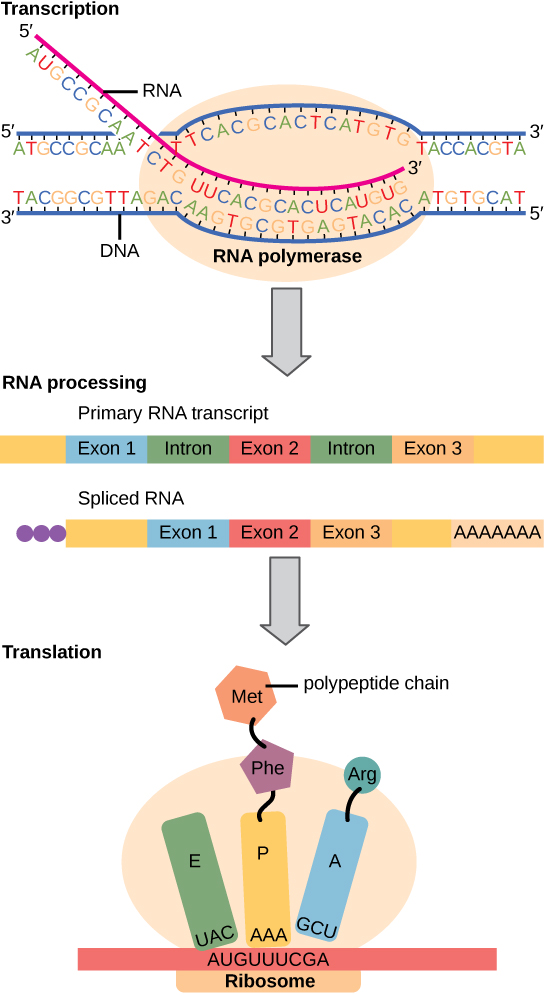
Figure 2: DNA to RNA, splicing, and translation to protein.
Transcript example
Transcript [1] link to ensemble/UBE2W-207
- Transcript UBE2W-207 ENST00000602593.6 8379bp 151aa.
- Exon 1 is 103 nucleotides.
- position start 73,878,910 end 73,878,808.
Transcript [2] link to ensemble/UBE2W-208
- Transcript UBE2W-208 ENST00000650817.1 8459bp 191aa.
- Exon 1 is 55 nucleotides.
- position start 73,878,862 end 73,878,808.
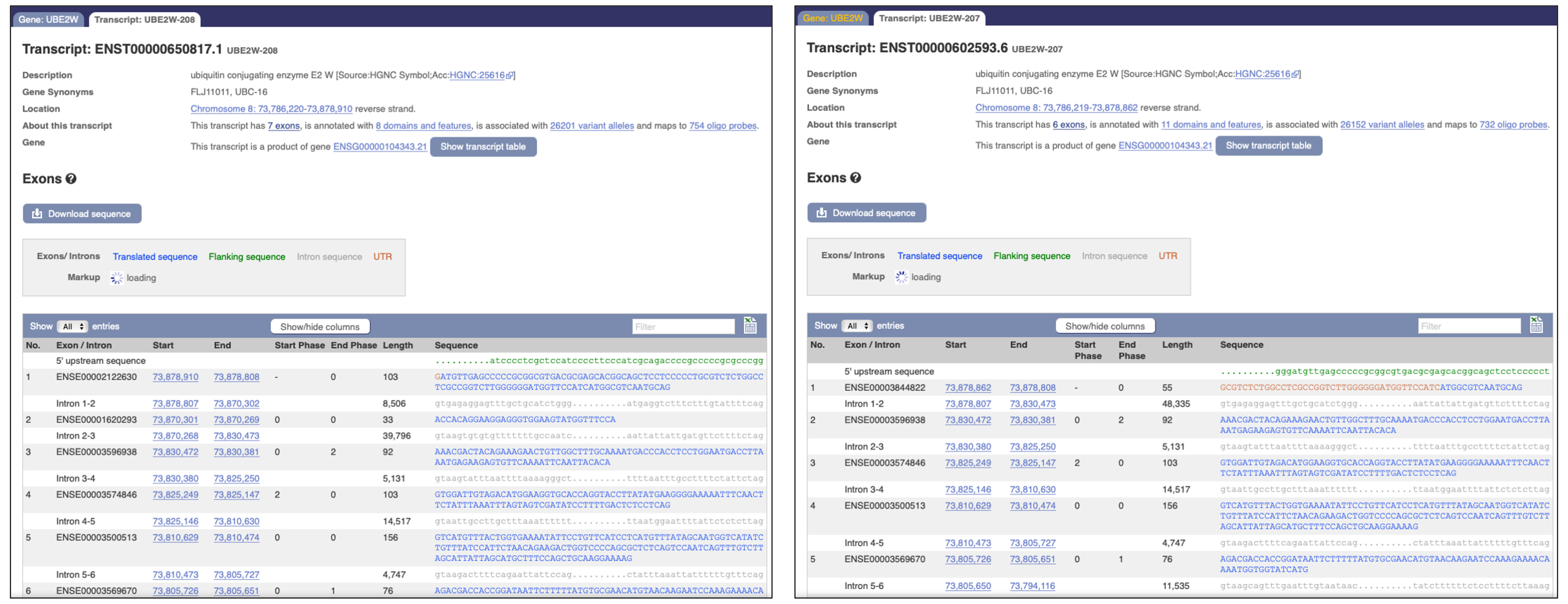
Figure 3: Screenshots from the Ensembl link listed above, showing two different transcripts based on the genomic DNA from one gene.
Mutation example
A DNA mutation at position g.73,878,863 A>T
This mutation will only affect transcript [2] at cDNA (mRNA) position c.2A>T and protein position p.Met1Leu.
Transcript [1] is not affected because the coding sequence begins later in the DNA sequence.
cDNA note
The cDNA position notation is used for simplicity. We assume the parts of genomic DNA that will remain after splicing (introns and upstream/downstream sequences removed), and call this the coding DNA (cDNA). We can physically create cDNA when we collect mRNA from a cell and do reverse Transcription PCR (RT-PCR) to make a DNA copy.
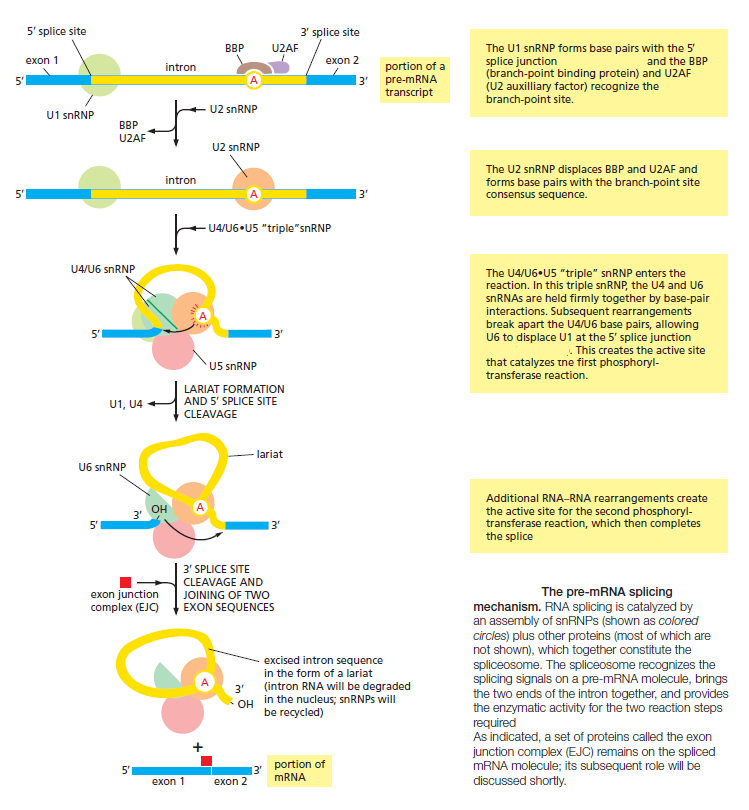
Figure 4: mRNA splicing in more detail.
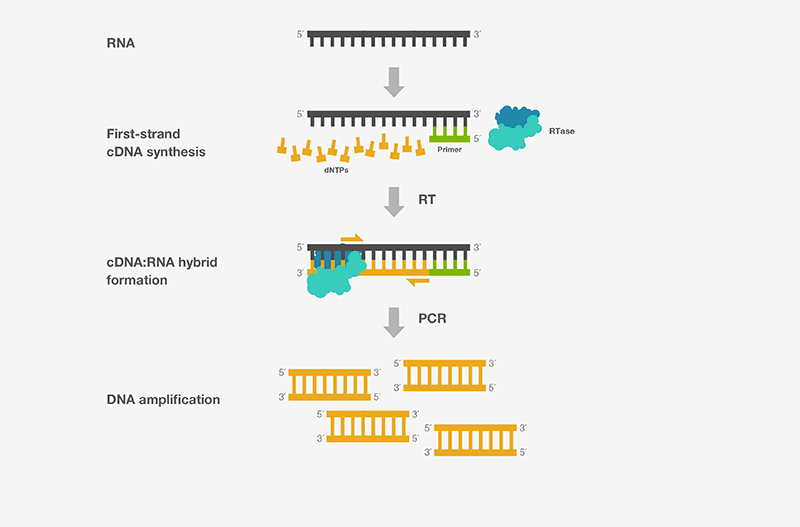
Figure 5. Simplified illustration of reverse transcription from an already-spliced RNA transcript to cDNA as used in the lab or performed by some (RNA genome) viruses.