Building the future
A collection of inspiration sources
To build the infrastructure of actionable clinical genomics and precision medicine, I am working on our automated results platforms. An important requirement is the translation of results to users, such as doctors and patients. This page hosts some examples of viz inspo that I draw from. Some pages or links may be dead - this repo is for design reference.
St. Jude cloud
The St. Jude-Washington University Pediatric Cancer Genome Project became the world’s most ambitious effort to discover the origins of childhood cancer and seek new cures. Here are a few pages to see:
- Homepage https://www.stjude.org/research.html
- Cloud apps https://www.stjude.cloud
- Protein paint https://pecan.stjude.cloud/proteinpaint/nras
- Analysis workflows https://platform.stjude.cloud/workflows
- Visualisations https://viz.stjude.cloud/visualizations

Figure: Examples from St. Jude cloud. Left: Analysis workflows each contain a one-page tutorial on running the app and include screenshots, code snippets, data types, etc. It serves as the user documentation. Top right: Apps include (i) the genomics platform - shown in previous panel - it documents the analysis workflows available on St. Jude. (ii) PeCan - shown in next panel. (iii) Visualisation community app (not shown) tracks the collection of all interactive or published viz such as HiC data, genome browsers, genomepaint splicing, etc. Bottom right: The PeCan app - interactive visualizations of variants and datasets used by all members.
 Figure: Great examples from St. Jude viz community. No comment or labels, just some examples.
Figure: Great examples from St. Jude viz community. No comment or labels, just some examples.
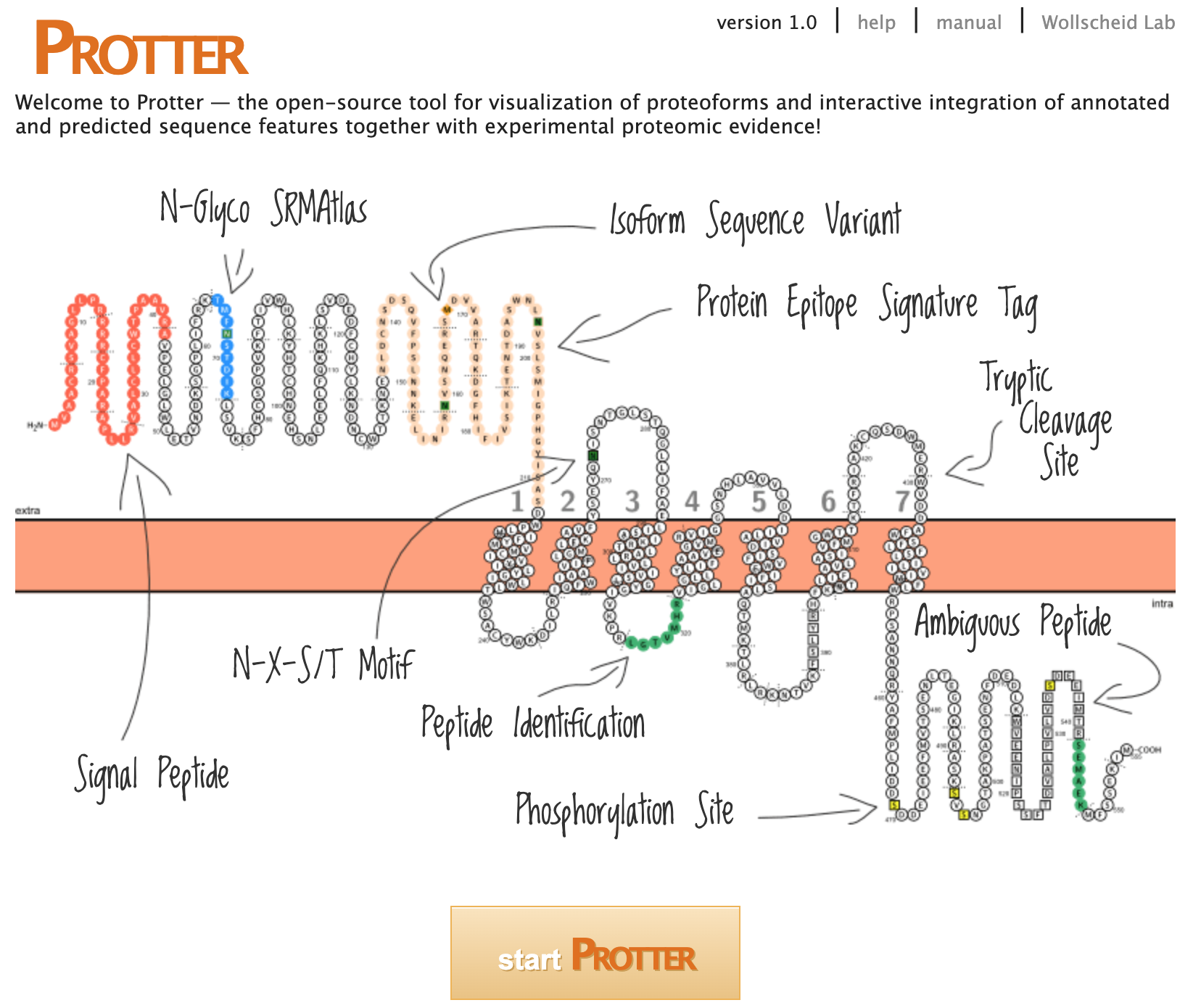
Protter
Protter: https://wlab.ethz.ch/protter/start/
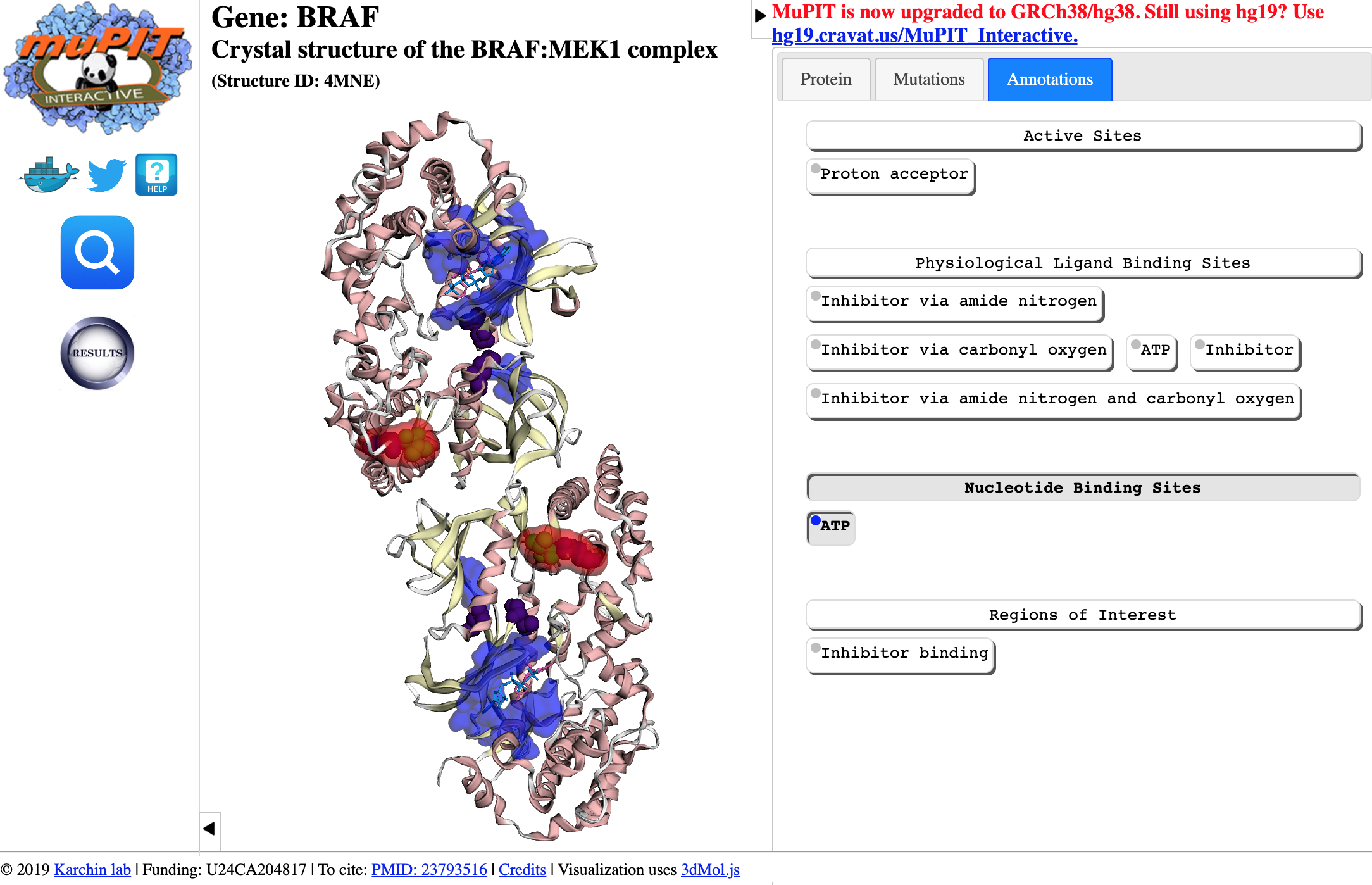
MuPIT
MuPIT: http://mupit.icm.jhu.edu/MuPIT_Interactive/
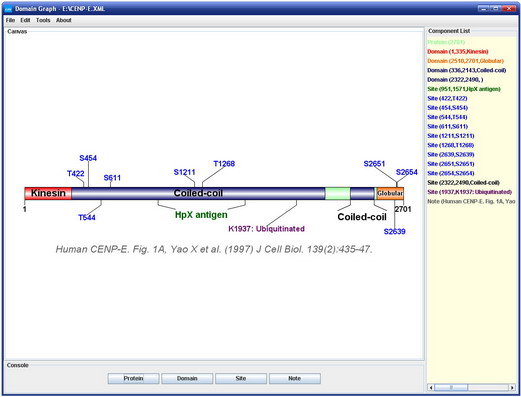
DOG
ggtranscript
ggtranscript: https://dzhang32.github.io/ggtranscript/