Pharmacogenomic stargazing
Note: Manual curation of gene-drug interaction is not recommended and therefore simplification of nomenclature is not advised. Relying on star allele labelling goes against the recommended HGVS sequence variant nomenclature. The following protocol for is provided as one step in a larger process of annotation in genomic analysis.
Stargazer genotyping
As reported in “Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program” (Taliun et al., 2021), use a method of known pharmacogene (PGx) variation annotation that can be adapted for many other drug-gene database applications. The star allele nomenclature method is applied to large scale data to screen for possible pharmacogenomic interactions. This consists of annotation with a simple notation for quick recognition of allelic variation in genes impacting drug metabolism, disposition and response (e.g. of interest to clinicians prescribing medications). The Pharmacogene Variation (PharmVar) consortium repository is used to label human cytochrome P450 (CYP) genes for known PGx variation.
Methods
Identification of CYP2D6 alleles using Stargazer’s genotyping pipeline. Details of the Stargazer genotyping pipeline have been described previously (Lee et al., 2019).
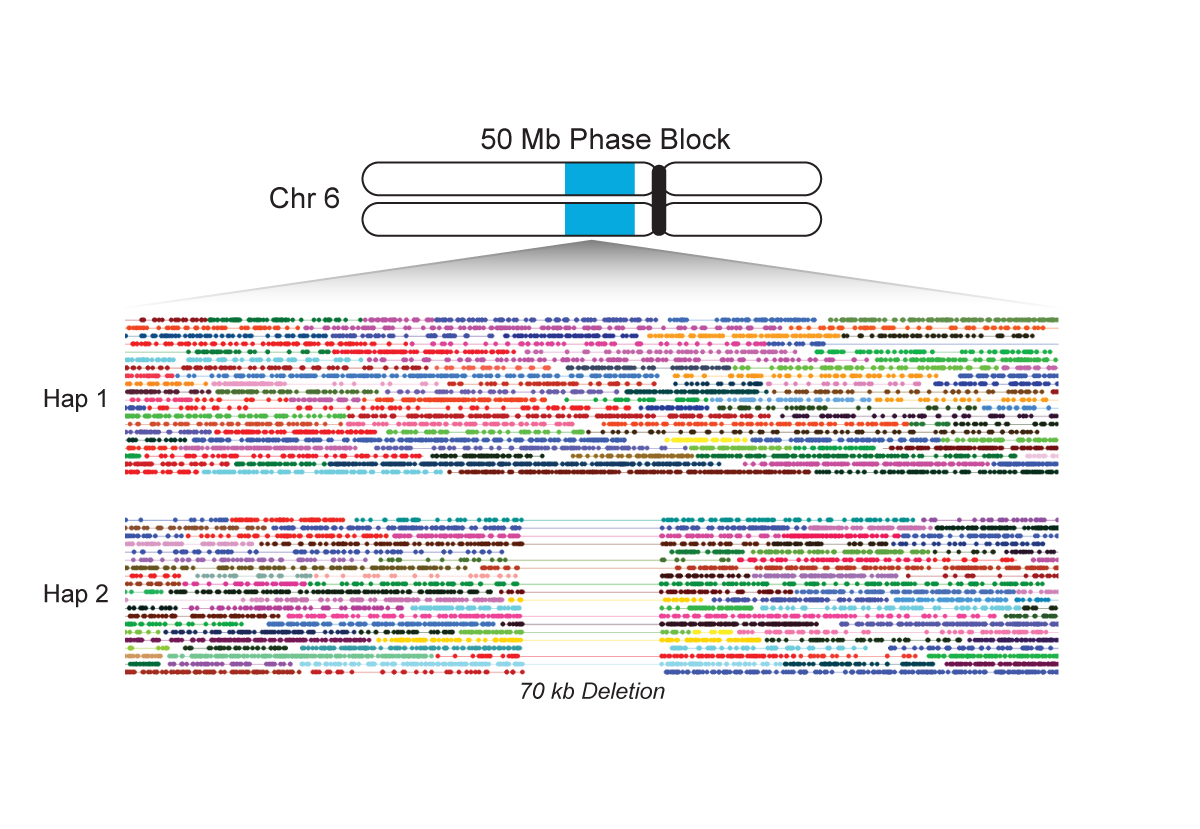
Background note: Haplotypes are group of alleles that are inherited together from a single parent. They are sequenced from individual DNA strands. These strands can be phased to reconstruct the inheritence pattern.
Phased haplotype (from 10Xgenomics.com):
- GATK-HaplotypeCaller
- SNVs and indels in were assessed from a VCF file generated using GATK-HaplotypeCaller (McKenna et al., 2010).
- Phase Beagle
- The VCF file was phased using the program Beagle (Browning & Browning, 2007) and the 1000 Genomes Project haplotype reference panel.
- Star alleles (described below)
- Phased SNVs and indels were then matched to star alleles.
In parallel,
- GATK-DepthOfCoverage
- Read depth was calculated from BAM files using GATK-DepthOfCoverage (McKenna et al., 2010).
- Copy number
- Read depth was converted to copy number by performing intra-sample normalization (Lee et al., 2019).
- Structural variants
- After normalization, structural variants were assessed by testing all possible pairwise combinations of pre-defined copy number profiles against the observed copy number profile of the sample.
- Changepoint
- For new SVs, breakpoints were statistically inferred using changepoint (Killick & Eckley, 2014).
- Output
- Information regarding new SVs was stored and used to identify subsequent SVs in copy number profiles.
- Output data included individual diplotypes, copy number plots and a VCF of SNVs and indels that were not used to define star alleles.
Stargazer summary
From Stargazer homepage - quote:
“Stargazer is a bioinformatics tool for calling star alleles (haplotypes) in PGx genes using data from NGS or SNP array. Stargazer can accept NGS data from both WGS and TS. “ Stargazer identifies star alleles by detecting SNVs, indels, and SVs. Stargazer can detect complex SVs including gene deletions, duplications, and hybrids by calculating paralog-specific copy number from read depth.”
Star alleles
From bredagenetics.com - quote:
“Genetic variants identifiable as pharmacogenomic markers are described by utilizing a special nomenclature, which is not elsewhere used in genetics. It is the so-called star allele nomenclature. In this nomenclature, alleles aren’t identified by their cDNA or genomic position (as it usually happens with all other genetic variants – see HGVS nomenclature), but through the means of numbers and letters, separated from the gene name by a star (star allele nomenclature). For example: CYP3A5*2 identifies the genetic variant in the CYP3A5 gene at the genomic position g.27289C>A, which leads to the amino acid substitution p.T398N. The star allele nomenclature is thought to be faster and easier for non-specialized professionals in identifying important pharmacogenetic alleles, helping them avoid transcription mistakes which may be more frequent by using the standard HGVS nomenclature.
“Alleles are marked with a star (*).
“A patient with ultrarapid drug metabolism harbors double or multiple copies of an allele with normal or increased functionality, whereas patients with intermediate or poor drug metabolism have one or more alleles with reduced functionality (these alleles are typically consistent with inactivating mutations or large gene deletions). The term extensive metabolizer is used instead to describe those individuals with two standard copies of the normally functional allele. Extensive metabolizers are therefore carrying the wild-type allele, also called consensus allele, which corresponds to the allele *1 in the star allele nomenclature. The numbers *2, *3, *4 and so on represent alleles with altered functionality which may lead to profiles of increased or reduced drug metabolism.
“By using one single star allele one can identify not just a single variant, but even a group of variants.”
A full list of cytochrome P450 (CYP) alleles with star notation can be found via the Pharmacogene Variation (PharmVar) consortium, a central repository for PGx variation that focuses on haplotype structure and allelic variation.
Published example
Taliun et al. (missing reference) (Zhou, 2009) (Crews et al., 2014). More than 150 CYP2D6 haplotypes have been described, some involving a gene conversion with its nearby non-functional but highly similar paralogue CYP2D7.
CYP2D6 interaction (from miro.medium.com):
They performed CYP2D6 haplotype analysis for all 53,831 TOPMed individuals (Lee et al., 2019) (Lee et al., 2019). Called a total of 99 alleles (66 known and 33 novel) representing:
- increased function,
- decreased function and
- loss of function (Supplementary Table 12).
Nineteen known alleles and all novel alleles were defined by structural variants, including complex CYP2D6-CYP2D7 hybrids and extensive copy number variation, which ranged from zero to eight gene copies (Supplementary Figs. 27, 28).
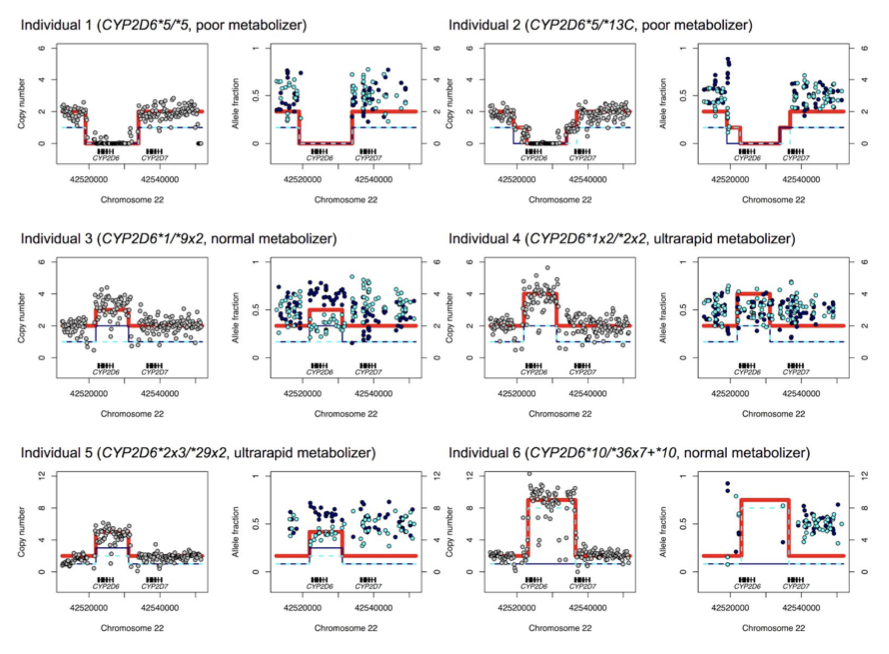
Supplementary Figure 27. Examples of CYP2D6 star alleles (haplotypes) with structural variation detected by the Stargazer program. Each panel displays Stargazer’s copy number profile (left) and allele fraction profile (right) for an individual sample (N=6). Also shown are CYP2D6 diplotypes and phenotype predictions from Stargazer. Gray dots indicate the sample’s per-base copy number estimates computed from read depth. The navy solid line and the cyan dashed line represent copy number profiles for each haplotype. The red line represents the copy number profile for both haplotypes combined. Navy dots and cyan dots indicate allele fraction estimates computed from allelic read depth for each haplotype. More examples can be found in the Database of Pharmacogenomic Structural Variants or DPSV \url{https://stargazer.gs.washington.edu/stargazerweb/res/dpsv.html}
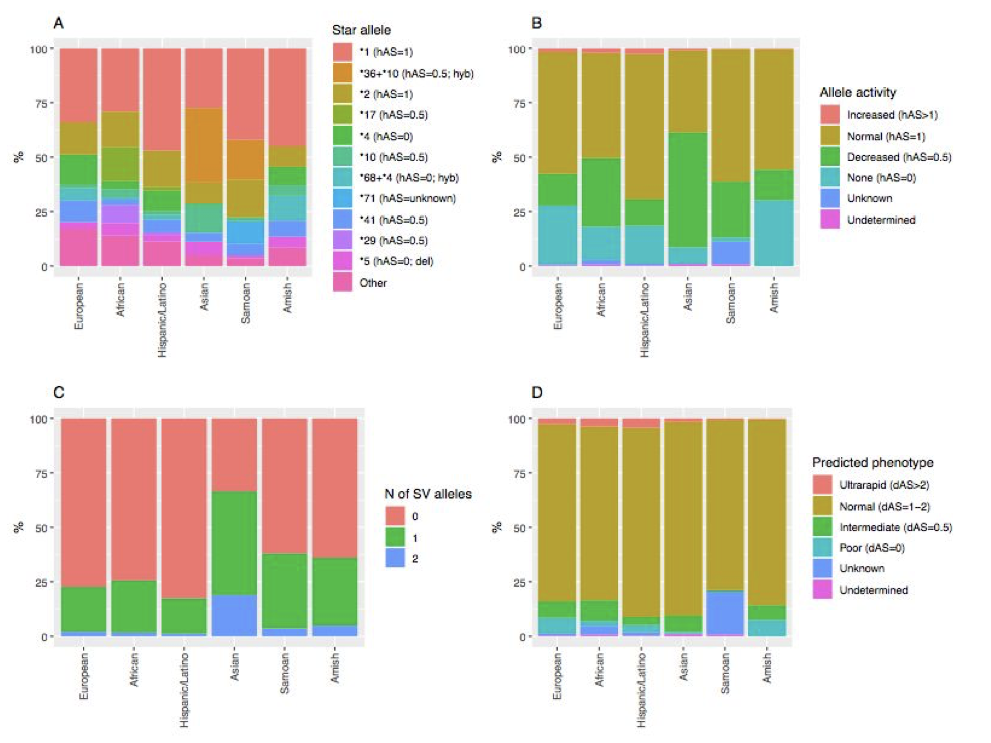
Supplementary Figure 28. Summary of CYP2D6 haplotype analysis using the Stargazer program. Population-specific frequencies for (A) common CYP2D6 star alleles, (B) haplotype activity, (C) SV-defined haplotypes, and (D) predicted metabolism phenotypes. Abbreviations: hAS, haplotype activity score; dAS, diplotype activity score; N, number; SV, structural variation; del, whole gene deletion; hyb, CYP2D6/CYP2D7 hybrid.
References
- Taliun, D., Harris, D. N., Kessler, M. D., Carlson, J., Szpiech, Z. A., Torres, R., Taliun, S. A. G., Corvelo, A., Gogarten, S. M., Kang, H. M., & others. (2021). Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature, 590(7845), 290–299.
- Lee, S.-been, Wheeler, M. M., Patterson, K., McGee, S., Dalton, R., Woodahl, E. L., Gaedigk, A., Thummel, K. E., & Nickerson, D. A. (2019). Stargazer: a software tool for calling star alleles from next-generation sequencing data using CYP2D6 as a model. Genetics in Medicine, 21(2), 361–372.
- McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., Garimella, K., Altshuler, D., Gabriel, S., Daly, M., & others. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research, 20(9), 1297–1303.
- Browning, S. R., & Browning, B. L. (2007). Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. The American Journal of Human Genetics, 81(5), 1084–1097.
- Killick, R., & Eckley, I. (2014). changepoint: An R package for changepoint analysis. Journal of Statistical Software, 58(3), 1–19.
- Zhou, S.-F. (2009). Polymorphism of human cytochrome P450 2D6 and its clinical significance. Clinical Pharmacokinetics, 48(12), 761–804.
- Crews, K. R., Gaedigk, A., Dunnenberger, H. M., Leeder, J. S., Klein, T. E., Caudle, K. E., Haidar, C. E., Shen, D. D., Callaghan, J. T., Sadhasivam, S., & others. (2014). Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clinical Pharmacology & Therapeutics, 95(4), 376–382.
- Lee, S.-been, Wheeler, M. M., Thummel, K. E., & Nickerson, D. A. (2019). Calling star alleles with stargazer in 28 pharmacogenes with whole genome sequences. Clinical Pharmacology & Therapeutics, 106(6), 1328–1337.